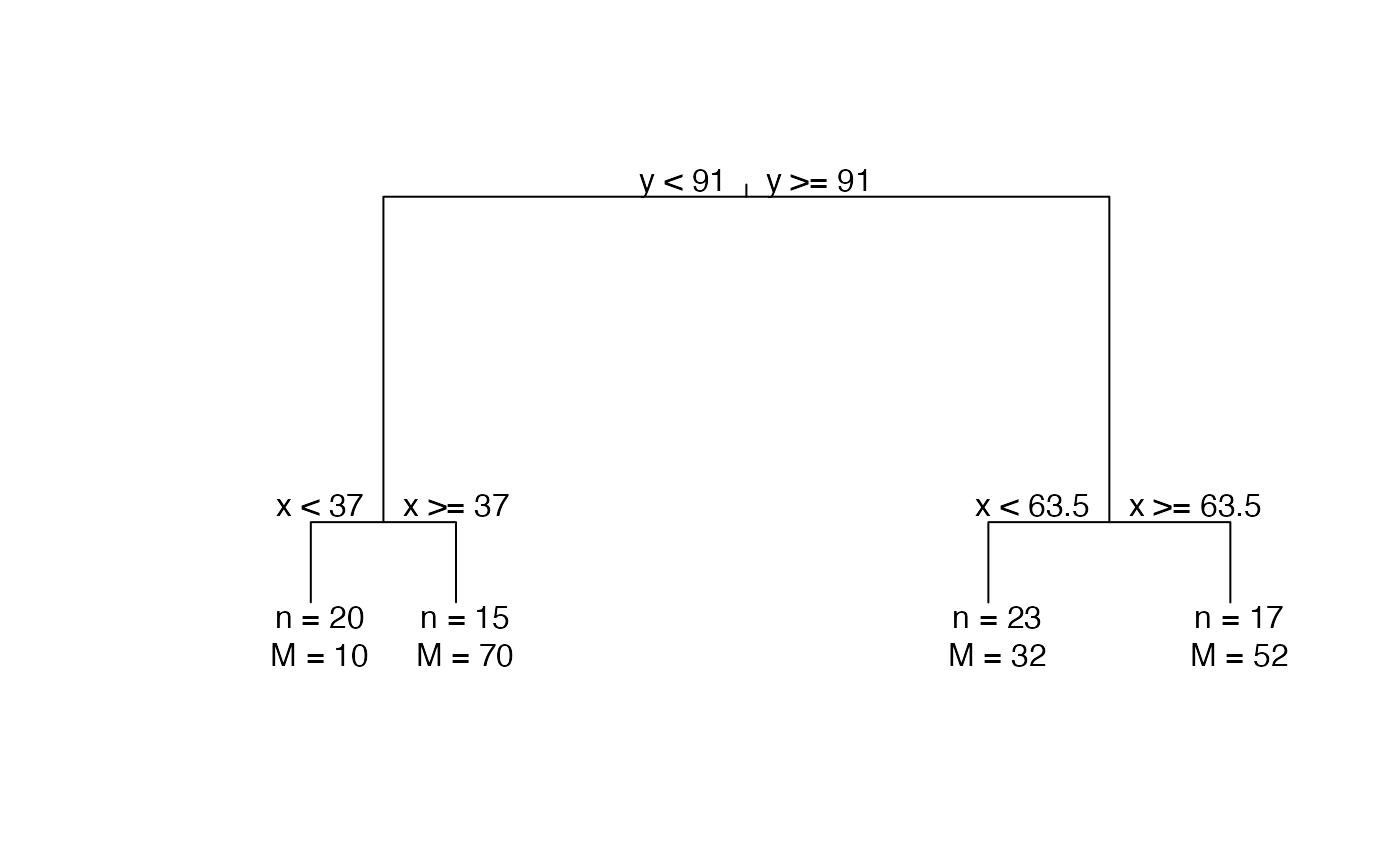
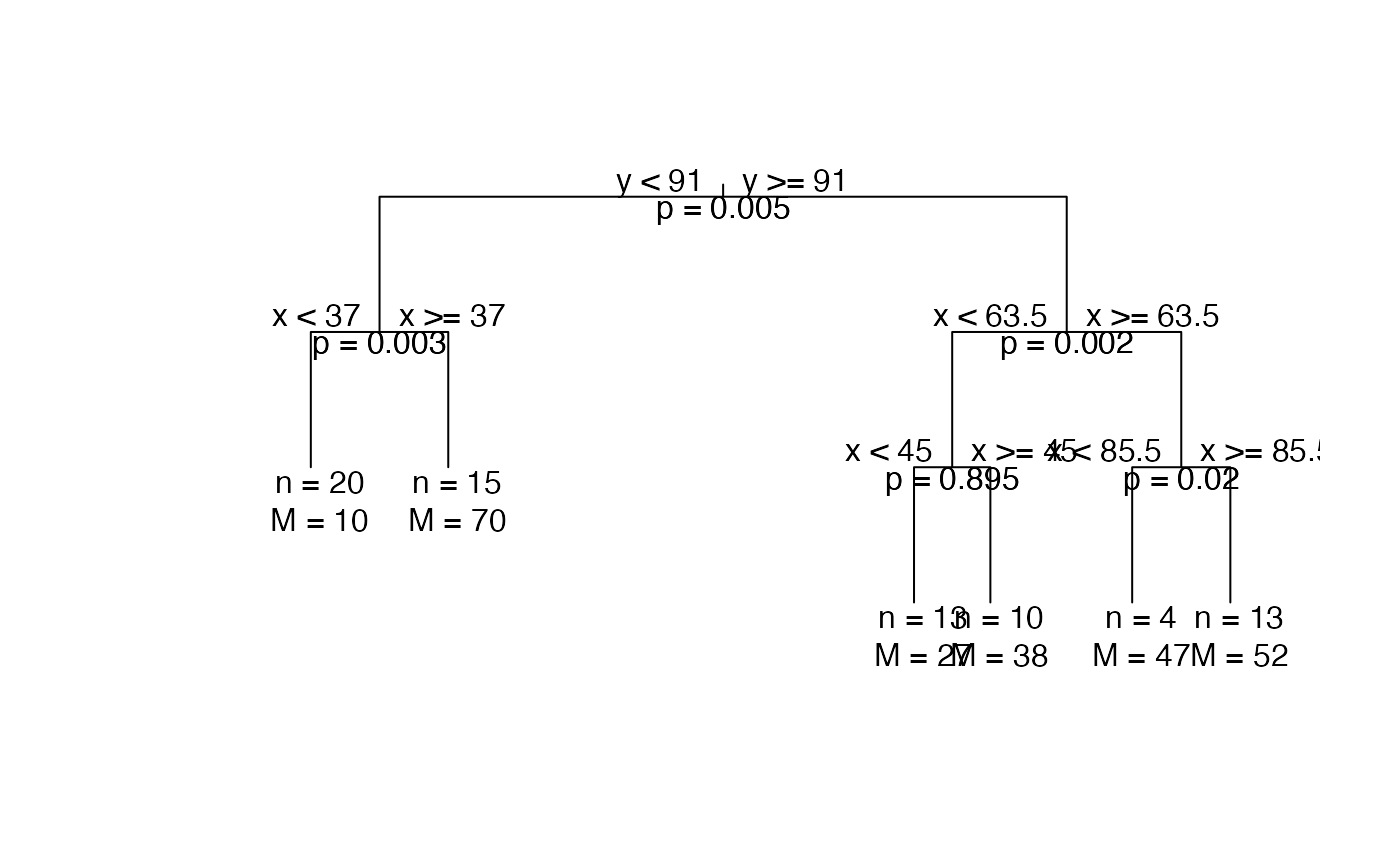
Print the MonoClust tree in the form of dendrogram.
# S3 method for MonoClust plot( x, uniform = FALSE, branch = 1, margin = c(0.12, 0.02, 0, 0.05), minbranch = 0.3, text = TRUE, which = 4, stats = TRUE, abbrev = c("no", "short", "abbreviate"), digits = getOption("digits") - 2, cols = NULL, col.type = c("l", "p", "b"), rel.loc.x = TRUE, show.pval = TRUE, ... )
Arguments
| x | MonoClust result object. |
|---|---|
| uniform | If TRUE, uniform vertical spacing of the nodes is used; this may be less cluttered when fitting a large plot onto a page. The default is to use a non-uniform spacing proportional to the inertia in the fit. |
| branch | Controls the shape of the branches from parent to child node. Any number from 0 to 1 is allowed. A value of 1 gives square shouldered branches, a value of 0 give V shaped branches, with other values being intermediate. |
| margin | An extra fraction of white space to leave around the borders of the tree. (Long labels sometimes get cut off by the default computation). |
| minbranch | Set the minimum length for a branch to |
| text | Whether to print the labels on the tree. |
| which | Labeling modes, which are:
|
| stats | Whether to show statistics (cluster sizes and medoid points) on the tree. |
| abbrev | Whether to print the abbreviated versions of variable names. Can be either "no" (default), "short", or "abbreviate". Short forms of them can also be used. If "no", the labels recorded in If "short", variable names will be turned into "V1", "V2", ... If "abbreviate", |
| digits | Number of significant digits to print. |
| cols | Whether to shown color bars at leaves or not. It helps matching
this tree plot with other plots whose cluster membership were colored. It
only works when |
| col.type | When |
| rel.loc.x | Whether to use the relative distance between clusters as x coordinate of the leaves. Default is TRUE. |
| show.pval | If MonoClust object has been run through |
| ... | Arguments to be passed to |
Value
A plot of splitting rule.
Examples
library(cluster) data(ruspini) # MonoClust tree ruspini4sol <- MonoClust(ruspini, nclusters = 4) plot(ruspini4sol)# \donttest{ # MonoClust tree after permutation test is run ruspini6sol <- MonoClust(ruspini, nclusters = 6) ruspini6_test <- perm.test(ruspini6sol, data = ruspini, method = "sw", rep = 1000) plot(ruspini6_test, branch = 1, uniform = TRUE)# }