Print the PULS tree in the form of dendrogram.
Arguments
- x
A
PULSobject.- branch
Controls the shape of the branches from parent to child node. Any number from 0 to 1 is allowed. A value of 1 gives square shouldered branches, a value of 0 give V shaped branches, with other values being intermediate.
- margin
An extra fraction of white space to leave around the borders of the tree. (Long labels sometimes get cut off by the default computation).
- text
Whether to print the labels on the tree.
- which
Labeling modes, which are:
1: only splitting variable names are shown, no splitting rules.
2: only splitting rules to the left branches are shown.
3: only splitting rules to the right branches are shown.
4 (default): splitting rules are shown on both sides of branches.
- digits
Number of significant digits to print.
- cols
Whether to shown color bars at leaves or not. It helps matching this tree plot with other plots whose cluster membership were colored. It only works when
textisTRUE. EitherNULL, a vector of one color, or a vector of colors matching the number of leaves.- col.type
When
colsis set, choose whether the color indicators are shown in a form of solid lines below the leaves ("l"), or big points ("p"), or both ("b").- ...
Arguments to be passed to
monoClust::plot.MonoClust().
Examples
# \donttest{
library(fda)
# Build a simple fd object from already smoothed smoothed_arctic
data(smoothed_arctic)
NBASIS <- 300
NORDER <- 4
y <- t(as.matrix(smoothed_arctic[, -1]))
splinebasis <- create.bspline.basis(rangeval = c(1, 365),
nbasis = NBASIS,
norder = NORDER)
fdParobj <- fdPar(fdobj = splinebasis,
Lfdobj = 2,
# No need for any more smoothing
lambda = .000001)
yfd <- smooth.basis(argvals = 1:365, y = y, fdParobj = fdParobj)
Jan <- c(1, 31); Feb <- c(31, 59); Mar <- c(59, 90)
Apr <- c(90, 120); May <- c(120, 151); Jun <- c(151, 181)
Jul <- c(181, 212); Aug <- c(212, 243); Sep <- c(243, 273)
Oct <- c(273, 304); Nov <- c(304, 334); Dec <- c(334, 365)
intervals <-
rbind(Jan, Feb, Mar, Apr, May, Jun, Jul, Aug, Sep, Oct, Nov, Dec)
PULS4_pam <- PULS(toclust.fd = yfd$fd, intervals = intervals,
nclusters = 4, method = "pam")
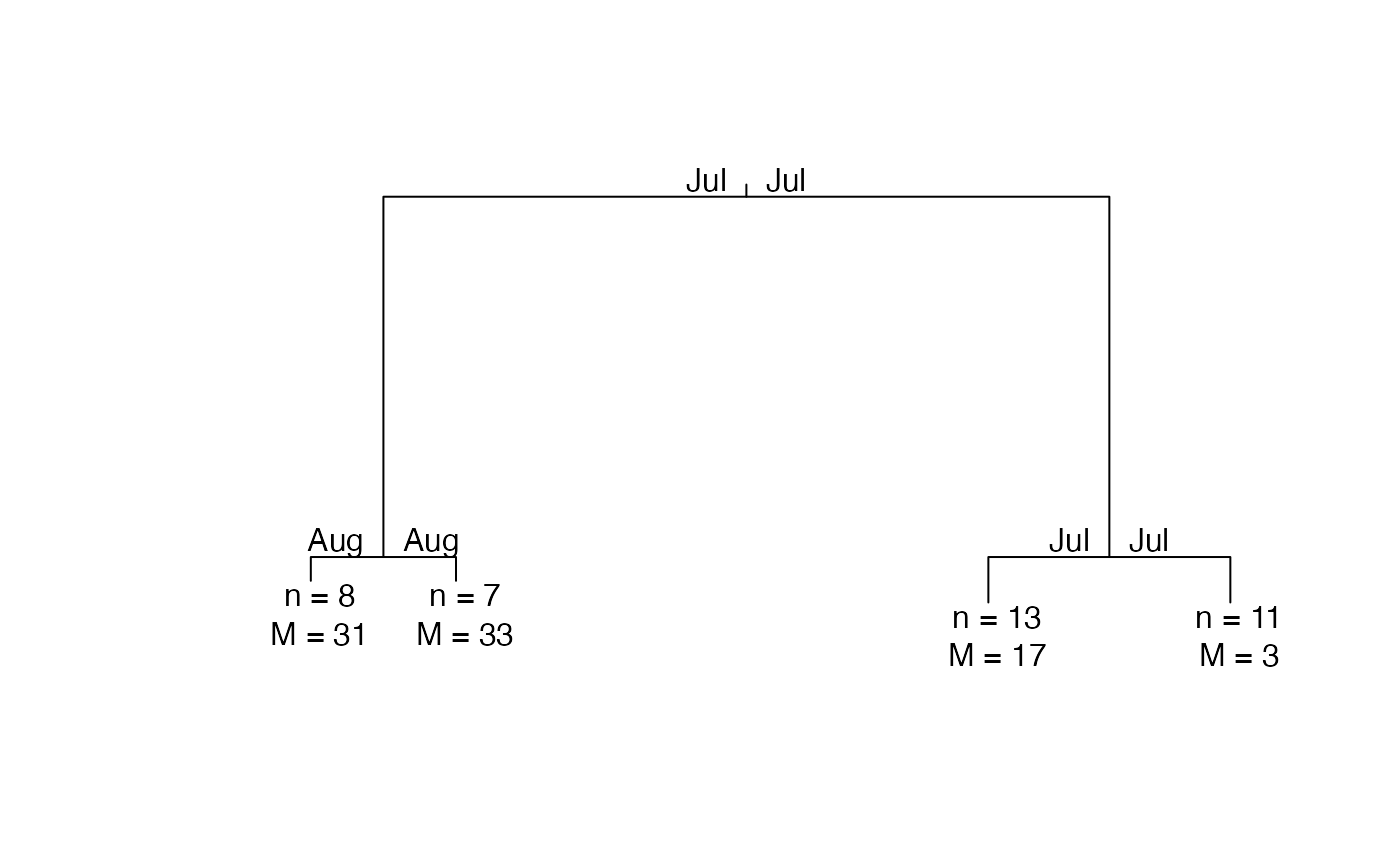
plot(PULS4_pam)
 # }
# }